Research
See my Google Scholar page for a full list of publications.
Probabilistic deconvolution of chromatin interaction signal in 3D genome data
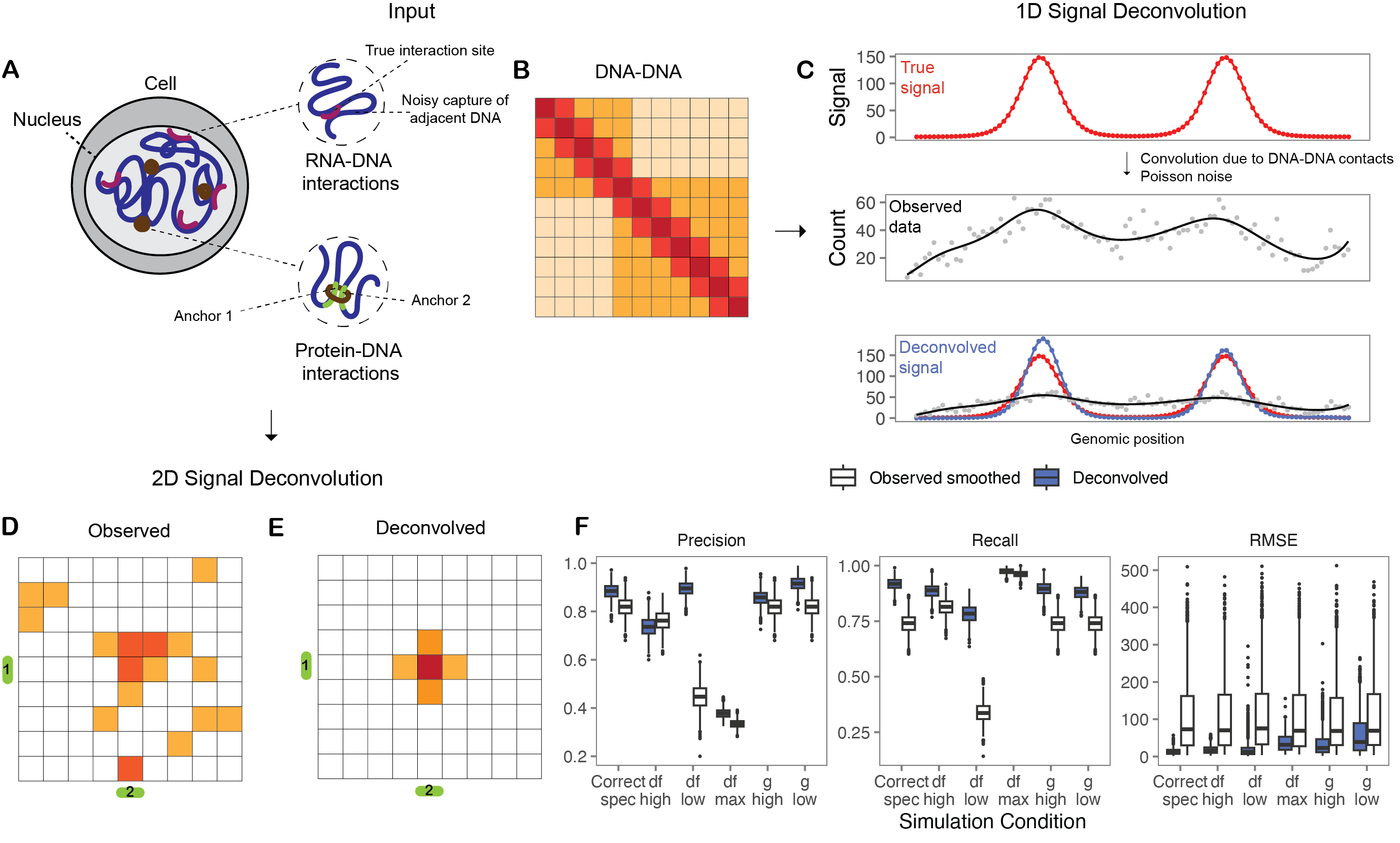
I developed a probabilistic model to describe the convolution of signal in 3D genome data such as HiChIP and RD-SPRITE. The method leverages DNA-DNA contacts to estimate the true, deconvolved signal.
Detection of allele-specific expression in spatial transcriptomics
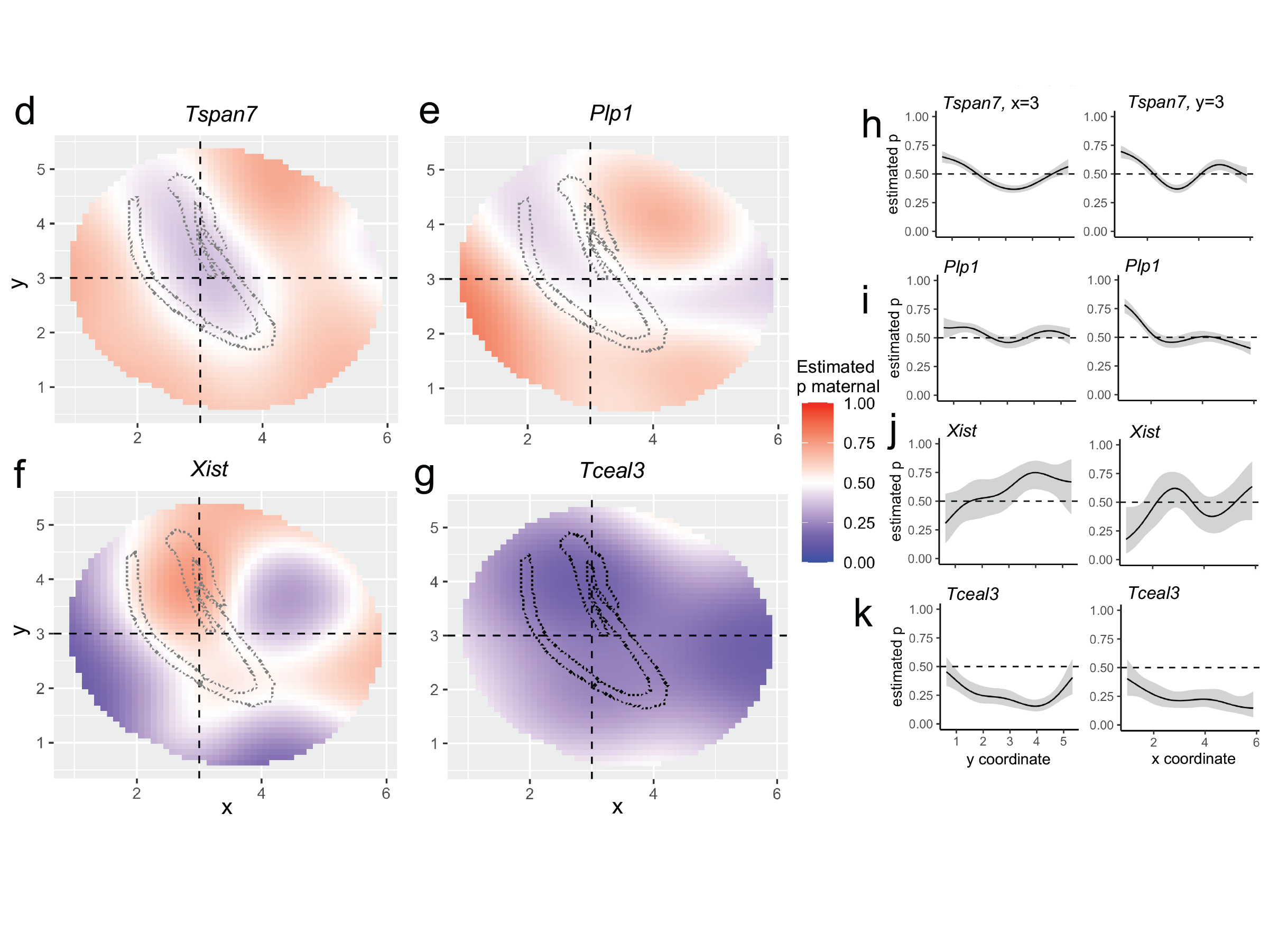
I developed a method to detect allele-specific expression in spatial transcriptomics. The method uses a beta-binomial likelihood and 2D smoothing splines to model spatial patterns.
